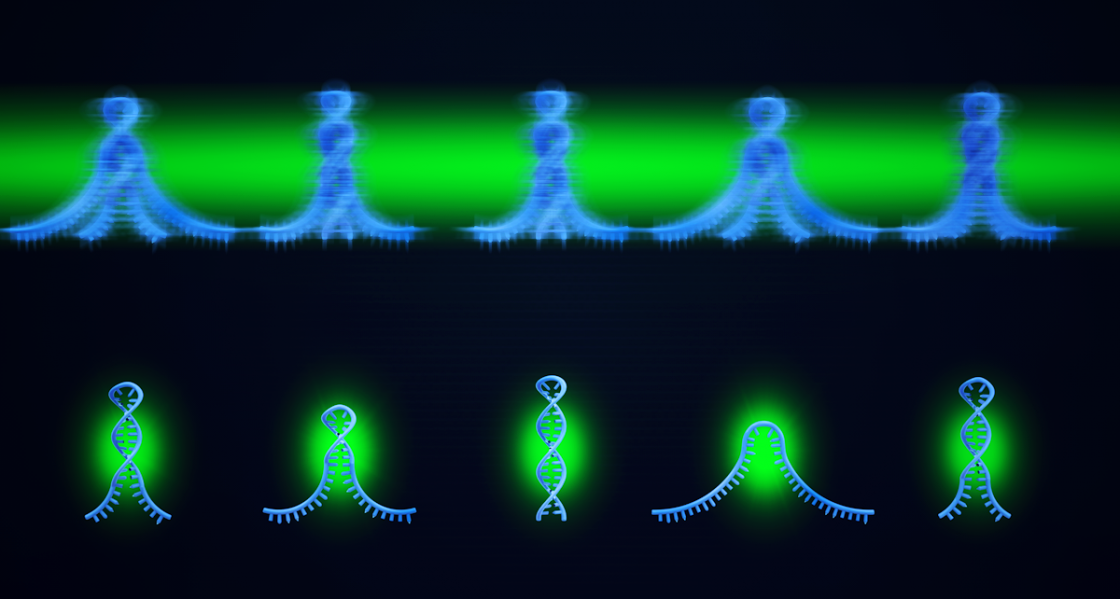
The basic question of how strands of nucleic acids (DNA and RNA) fold and hybridize has been studied thoroughly by biophysicists around the globe. In particular, there can be unexpected challenges in obtaining accurate kinetic data when studying the physics of how DNA and RNA fold and unfold at the single molecule level. One problem comes from temporal camera blur, as the cameras used to capture signal photons emitted by these molecules do so in a finite time window that can blur the image and thereby skew the kinetic data In a paper published in the Journal of Physical Chemistry B, JILA Fellow David Nesbitt, and first author, and graduate student, David Nicholson, propose an extremely simple yet broadly effective way to overcome this camera blur. According to Nicholson: "We wanted to measure the speed of nucleic acid folding, but to our dismay, we encountered a systematic bias that comes up when you do these kinetic measurements if you're not careful. Specifically, you get into trouble if the DNA is folding as quickly as the camera is recording images. So, we started thinking, how can we come up with a way to fix this problem?" In looking at their data, Nicholson and Nesbitt realized that they could reduce this systematic error and extend the domain of kinetic study in a surprisingly simple way by shining a light on the problem, in particular, a strobe light.
Why Throwing Data Away is Beneficial... Sometimes
Nicholson and Nesbitt realized that they could reduce the camera blur by shining pulsed, rather than continuous, laser light onto the molecules, reducing the time fraction of observation (i.e., duty cycle), but making their total error due to camera blur significantly lower. Nesbitt explained: "This was entirely David Nicholson’s idea, for which I simply gave him a little encouragement to pursue. The key idea is really like a stroboscope in a crowded night club or disco. Basically, we're flashing light onto the molecular 'dance' in a short time window faster than the camera frame rate. This of course requires us to throw away information between pulses, but at the same time, provides much better kinetic information from each pulse." The data thrown away can be accounted for by a mathematical correction, resulting in kinetics that are accurate even up to the camera frame acquisition rate, an order of magnitude in improvement. Nesbitt clarified the fix in more technical terms: “The method reduces the error because normal analysis of blurred objects has a built-in mathematical bias that tends to make kinetic analysis of these actions appear systematically slower.” Nicholson and Nesbitt had just found a simple solution for their problem in a stroboscope.
A stroboscope is a fancy word for a strobe light. Stroboscopic imaging, a process wherein an objects movement is represented by short light samples, has been used for many decades, though not in single-molecule kinetic measurements. "Actually, we thought for sure someone would have already published this concept long ago," Nicholson commented, "but after digging through the literature, it turns out there was still a lot of confusion about how to deal with single-molecule camera blur. So, we said, 'OK, this seems like something researchers could find useful.’" With regard to the simplicity of such a method, both Nicholson and Nesbitt initially wondered if this approach was even worth publishing at all. "Once you appreciate the basic idea behind the method, it seems so completely simple, and frankly, a bit obvious! We wondered,' is this really new and worth publishing?' But in fact, it's exactly those sorts of discoveries that belong in the literature because everyone can so easily implement it." The fact that this simple method had not already been published made Nicholson and Nesbitt more interested in making their method public for everyone to use. As Nicholson noted, this method would be inexpensive for researchers to implement, and they would see an immediate improvement in their range of measurement by as much as an order of magnitude. The team hoped that their method could save researchers time and effort when it came to fixing kinetic measurement bandwidth problems associated with camera blur. They also realized that this method would help save researchers valuable time when it came to data analysis and to extending their kinetic measurements up to the camera frame acquisition rate limit. Said Nesbitt: "I think comparing our stroboscopic method to a flash on a camera is helpful. We're taking images with this method to get rid of motion blur. It's allowing us to see our DNA molecules more crisply."
Spreading the Word
Nesbitt and Nicholson look forward to seeing their work implemented by other researchers, "The wonderful corollary is that, as camera technologies get better and faster, David Nicholson’s method should improve right along with them," Nesbitt explained. The benefits of this new method are not only cost-effective and easy to use, but clearly can adapt as the technology itself improves in the coming years.
Written by Kenna Castleberry, JILA Science Communicator


 The Physics Frontiers Centers (PFC) program supports university-based centers and institutes where the collective efforts of a larger group of individuals can enable transformational advances in the most promising research areas. The program is designed to foster major breakthroughs at the intellectual frontiers of physics by providing needed resources such as combinations of talents, skills, disciplines, and/or specialized infrastructure, not usually available to individual investigators or small groups, in an environment in which the collective efforts of the larger group can be shown to be seminal to promoting significant progress in the science and the education of students. PFCs also include creative, substantive activities aimed at enhancing education, broadening participation of traditionally underrepresented groups, and outreach to the scientific community and general public.
The Physics Frontiers Centers (PFC) program supports university-based centers and institutes where the collective efforts of a larger group of individuals can enable transformational advances in the most promising research areas. The program is designed to foster major breakthroughs at the intellectual frontiers of physics by providing needed resources such as combinations of talents, skills, disciplines, and/or specialized infrastructure, not usually available to individual investigators or small groups, in an environment in which the collective efforts of the larger group can be shown to be seminal to promoting significant progress in the science and the education of students. PFCs also include creative, substantive activities aimed at enhancing education, broadening participation of traditionally underrepresented groups, and outreach to the scientific community and general public.