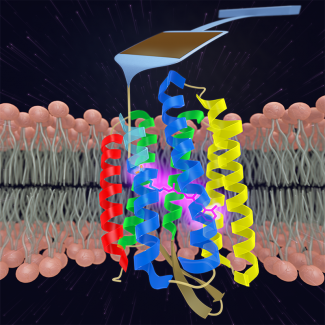
We are studying the energetics that stabilize membrane proteins by folding and unfolding individual molecules using AFM. Through such studies, we are making quantitative measurements of the energy barriers between obligatory, non-obligatory, and off-pathway intermediate states.
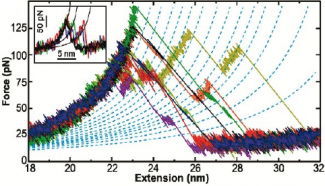
In our first work in this area, we used cantilevers optimized for 1-µs resolution to re-examine the unfolding of individual bacteriorhodopsin (bR) molecules (in native lipid bilayer) with a 100-fold higher time resolution and a 10-fold higher force precision than previous studies. We measured bR’s unfolding pathway with unprecedented detail, identifying structural elements associated with bR unfolding that were as small as two amino acids. Moreover, we resolved a longstanding discrepancy between AFM data and molecular dynamics over the fundamental size scale of bR’s unfolding: prior AFM work showed 2 intermediates (figure inset) while molecular dynamics predicted 10. Our enhanced assay revealed 14 intermediates in the unfolding of a pair of transmembrane helices.
To go beyond this tantalizing initial work, we developed a scheme to form a covalent bond in situ between bR and a PEG-coated AFM tip, rather than relying on non-specific tip-sample adhesion. Using this technology, we precisely characterized the first amino acids of bR that unfold when force is applied, a region obscured by tip-sample adhesion artifacts in prior measurements. In doing so, we uncovered a new unfolding intermediate that had gone undetected over the prior 18 years, despite being the most mechanically robust state in bR’s unfolding pathway (due to stabilization by the retinal cofactor) and occurring in 100% of unfolding records.
Identification of this stable unfolding intermediate, and of two reversible near-equilibrium unfolding transitions preceding it, enabled us to quantify the equilibrium (un)folding free energy ΔG0 of an eight-amino-acid region starting from the fully folded state of bR. Analysis of equilibrium and nonequilibrium data yielded consistent, high-precision determinations of ΔG0 via multiple techniques (force-dependent kinetics, Crooks fluctuation theorem, and inverse Boltzmann analysis). Importantly, our determination of ΔG0, and all of our measurements on bR, are performed in the protein’s native bilayer, an advance over traditional chemical-denaturation measurements performed in nonphysiological detergent micelles.
Excitingly, these technical advances to measure ΔG0 now allows us to use single-molecule unfolding experiments to access quantities of direct interest in understanding membrane protein folding and dynamics. For example, by making measurements in wild-type and mutant bR, we have determined ΔΔG for select point mutants, thereby establishing a platform for determining ΔΔG for a fully folded membrane protein embedded in its native bilayer.


 The Physics Frontiers Centers (PFC) program supports university-based centers and institutes where the collective efforts of a larger group of individuals can enable transformational advances in the most promising research areas. The program is designed to foster major breakthroughs at the intellectual frontiers of physics by providing needed resources such as combinations of talents, skills, disciplines, and/or specialized infrastructure, not usually available to individual investigators or small groups, in an environment in which the collective efforts of the larger group can be shown to be seminal to promoting significant progress in the science and the education of students. PFCs also include creative, substantive activities aimed at enhancing education, broadening participation of traditionally underrepresented groups, and outreach to the scientific community and general public.
The Physics Frontiers Centers (PFC) program supports university-based centers and institutes where the collective efforts of a larger group of individuals can enable transformational advances in the most promising research areas. The program is designed to foster major breakthroughs at the intellectual frontiers of physics by providing needed resources such as combinations of talents, skills, disciplines, and/or specialized infrastructure, not usually available to individual investigators or small groups, in an environment in which the collective efforts of the larger group can be shown to be seminal to promoting significant progress in the science and the education of students. PFCs also include creative, substantive activities aimed at enhancing education, broadening participation of traditionally underrepresented groups, and outreach to the scientific community and general public.